
A Lynx With a Legacy
A Lynx With a Legacy
6 minute read
The Yukon Wildlife Preserve is deeply saddened to announce the passing of its beloved male lynx on Saturday, November 9th. This exceptional animal, who joined the Preserve’s collection in 2010, was a symbol of resilience and scientific importance, leaving behind a legacy that will not be forgotten.

Photo credit: L.Caskenette 2018
Born in the wild in 2008, the male lynx’s early life was marked by an unfortunate accident during a research project in the Edmonton, Alberta area. The injury required the amputation of his back left leg, a traumatic event that ultimately led to his rescue and relocation to the Yukon Wildlife Preserve. For the first eight years of his time at the Preserve, he remained behind the scenes, away from the public eye, while another male lynx, who had been part of the collection, cohabited with the other lynx.

Photo credit: Jake Paleczny
The passing of this other male lynx, who lived to almost 21 years of age, in December 2017, allowed for the three-legged lynx to join the female lynx, and he quickly became a well-known and cherished resident of the Preserve. His unique physical appearance, the result of his injury, made him stand out as a symbol of adaptability and strength.

Photo credit: Jake Paleczny Male lynz born in 1997. Died in December of 2017.
But the three-legged lynx was not only special because of his physical traits—his genetics were also extraordinary. His wild origins made him an invaluable member of the Species Survival Plan (SSP) program, where he was recognized for contributing genetically to the broader conservation effort for Canada lynx. As part of an ambitious scientific study in 2018, the Yukon Wildlife Preserve facilitated the mapping of this lynx’s genome in collaboration with the University of Toronto. This groundbreaking research provided insights into chromosome structure in mammals and offered important data for comparing the Canada lynx to the highly endangered Iberian lynx, helping to resolve scientific controversies surrounding the phylogenetic classification of this cat species.

Summer 2024. Photo credit: Ami Vitale National Geographic
Despite his distinct genetic heritage, the lynx’s time at the Preserve also had challenges. Although he made several attempts to breed with both the 2009 and 2014 female lynx, no successful litters were produced. Tragically, the one litter born to the older female lynx in 2021 did not survive. Despite these setbacks, the male lynx’s contributions to both the Preserve’s mission and scientific research were invaluable.

Summer 2024. Photo credit: Ami Vitale National Geographic
The loss of this remarkable animal is a bittersweet moment for the team at the Yukon Wildlife Preserve. His resilience, both in the face of adversity and throughout his life, will be remembered by all who had the privilege of caring for him.
Taken November 5 2024. Photo credit: L.Caskenette
The Yukon Wildlife Preserve remains committed to preserving and protecting wildlife, and will continue to honor the legacy of this exceptional lynx as part of its broader conservation efforts. As we learn more from the necropsy details we will share.
• • •
In 2017/2018 The Centre for Applied Genomics (TCAG), at The Hospital for Sick Children, and University of Toronto connected with the Preserve to ask for support to study chromosome structures in animals. As apart of the Canada 150 celebrations as well the genome mapping of several iconic Canadian speciesm including first a beaver, were completed. A blood sample was provided by our three legged lynx.
The karyotype (chromosome complement) of this resident Canada Lynx, was determined to be 38, XY (see picture). Having 38 chromosomes of the ascribed size is identical to that found in the domestic cat, indicating close relationship. The presence of a X- and a Y-chromosome confirmed the sex of the animal, as a male.
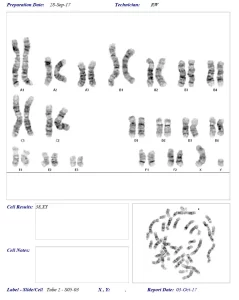
Background
The Centre for Applied Genomics (TCAG), at SickKids is headed by Stephen Scherer who is a recognized expert in the field of neurogenomics. Neurogenomics combines classical neurobiology (study of the brain) with the study of the genome (our genetic material) in the context of disease, such as Autism. The human genome is represented by a string of 4 chemical bases, over 3 billion bases long that is found in each cell of the body. The precise order or the sequence of these four chemical bases constitutes the organism’s genome, which defines who and what the organism is, whether it is a cat, a bird, or a tree etc. Essentially, the genome is the organism’s blueprint that is inherited to future generations. It is not hard to imagine that changes in the order of these bases in the genome could give rise to disease. In fact, most cancers are due to such changes, either due to plain bad luck or from exposure to chemicals or radiation and the like.
The technology exists to determine the “sequence” of the genome to give a heads up on disease or treatment. Scientists are in the era of the “$1,000 genome” a term coined by the marketing people. SickKids is the largest genome sequencing centre in Canada and they “sequence” about 10,000 patient genomes a year. You would recall the Human Genome is 3 billion bases long. But in order to produce a genome for only $1,000, the sequencing machine reproduces a patient’s genome as tiny low cost pieces, 400 million of them in fact, with each piece only about 300 bases long. These highly fragmented pieces of the genome are useful only because we already sort of know what the human genome looks like. These pieces are mapped individually onto the official “Human Reference Genome” and the differences between the two sequences (i.e. bases that are different, missing, or extra bases inserted when compared to the mapped position on the Human Reference Genome) are tabulated to produce a list of “mutations” for each patient. The current (2017 reference) official “Human Reference Genome” took nearly 15 years to build from scratch (from little pieces) at a cost of more than $US 3 billion dollars, paid for by the US government and various philanthropic organizations. This reference-based mapping method for genome sequencing works, and many human disease-causing mutations have been identified in this way.
So what is the problem?
The problem is that sequencing a patient’s genome by mapping and comparing short segments against the Human Reference Genome will not identify all the mutations. Many regions of a patient’s genome could be so different from the Reference Genome such that many short pieces derived from patient would not map at all; hence, large segments of the patient’s genome are not amenable to analysis. They estimate about 50% of mutations are missed. To get at all the mutations, they would need to move away from the current mapping approach and reconstruct the patient’s entire genome from the little pieces from scratch. This is process is called “de novo sequencing” (new or from scratch) and is similar to how the original Human Reference Genome was constructed. The trick is that, unlike the creation of the Human Reference Genome, we cannot take 15 years and $3 billion dollars to assemble the genome of each patient.
So what are we doing?
We are now working on methods to perform de novo sequencing and genome assembly there are fast and cheap. To perfect de novo sequencing in an unbiased manner, we used a genome that previously had not been sequenced before. This was one of the reasons why we sequenced and assembled the Canadian Beaver Genome early this year. Other reasons, but just as important: it was Canada’s 150 anniversary and the beaver was not only named after Canada (Castor canadensis), but it had played an important role in the founding of this country; it was great for educational and environmental outreach; and finally, we want to beat an American group who wished to sequence the genome of their beaver football team mascot. At the end, we decisively won the Beaver Genome Race and our paper was published in Feb this year (See attached). We even made the cover of the journal, and had great response form the media.
Why the Lynx?
Despite the good progress made on the de novo sequencing of the beaver genome, the process is still too expensive for routine use at the hospital. We have new ideas on how to make the process better and cheaper. Hence, we need another un-sequenced genome to test the system. Moreover, the beaver genome was so well received; we decided to perform de novo sequencing and assembly of up to ten or more notable animal, bird, fish or plant in the next couple of years. The actual number we would do would depend on how low we can drive the price down. The short list of candidates include the organisms that are named after Canada (there are over eighty of them), have notable ties with Canada, or have compelling scientific rationale.
This is where the Canada Lynx (Lynx canadensis) comes in. No one had sequenced a Canada Lynx so far. From a scientific perspective, a Canada Lynx genome sequence would address a lot of the controversies relating to the phylogenetic assignments of this branch of the cat family, and it would also allow a detail comparison with the recent published genome of the highly endangered Ilberian lynx. Finally, the lynx has considerable public appeal. To push the technical envelope, we aim to sequence and assemble the Lynx genome at less half the cost of beaver genome whilst keeping the quality at the same level. Eventually, we would arrive at an efficient and cost-effective process for clinical use.
The lynx and hare were perfect Canadian canditaed to kickstart the project.

Lindsay Caskenette
Manager Visitor Services
Lindsay joined the Wildlife Preserve team March 2014. Originally from Ontario, she came to the Yukon in search of new adventures and new career challenges. Lindsay holds a degree in Environmental Studies with honours from Wilfrid Laurier University and brings with her a strong passion for sharing what nature, animals, and the environment can teach us.





























